Predicting#
mbGDML was initially designed just for GDML models, hence the name.
However, other ML potentials or even QC methods can be used to make many-body predictions.
We designed an extensible, modular framework for many-body predictions using mbePredict.
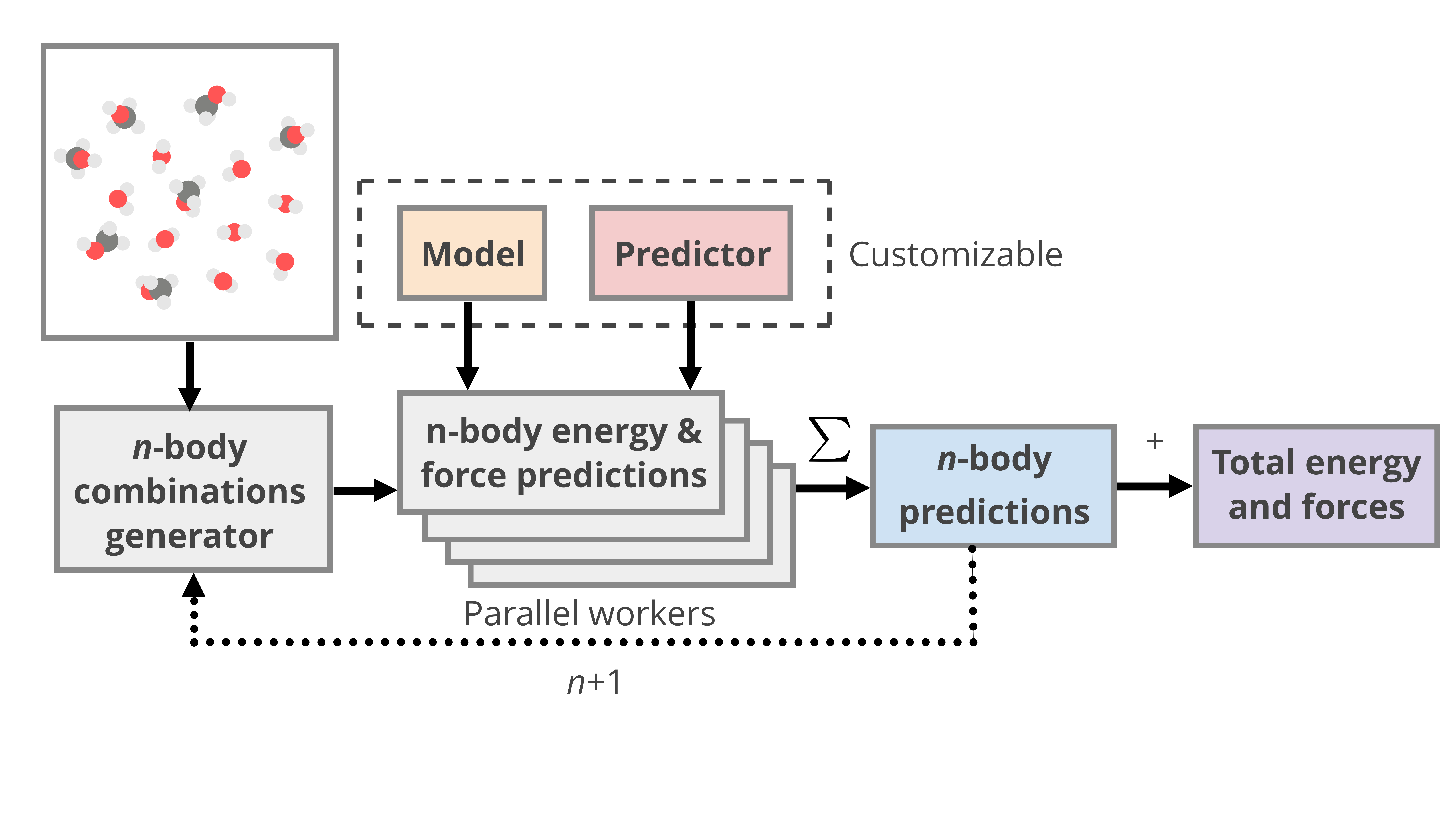
mbePredict requires \(n\)-body models and predictor functions.
Model#
Information needed to make quantum chemistry or machine learning predictions are contained in objects that inherit the Model class.
For example, gdmlModel stores GDML parameters for the predict function.
Predict#
Predict functions perform the actual predictions from the respective model object along with
Z: atomic numbers,R: cartesian coordinates of a single structure,entity_ids: integers specifying atoms that make up individual entities or fragments,entity_combs: unique combinations of entity IDs to predict.
For example, predict_gdml() will sum up all \(n\)-body combinations for their contribution to R’s energy and forces.
You can also have decomposed predict functions (i.e., predict_gdml_decomp()) that return all individual \(n\)-body energies and forces.
mbePredict#
mbePredict handles all aspects of making many-body expansion predictions with ML potentials.
Once initialized, predictions are made with the predict() method where we specify our system.
compute_nbody() is then called for each loaded model until all possible \(n\)-body contributions are accounted for.
Warning
Predictions are made by iterating through models and accounting for all compatible entities. If the model component IDs are different, no entity combinations are used.
Parallel predictions#
Many-body expansions are known for their curse of dimensionality: as the supersystem grows in size, so does the number of entity combinations.
These can be easily parallelized by assigning workers with specified batch sizes.
Currently, only a ray implementation for GDML is provided by specifying use_ray to True and setting n_workers.
Supported potentials#
mbGDML already provides support for the following potentials:
Gradient-Domain Machine Learning (GDML) with
gdmlModelandpredict_gdml()Gaussian Approximation Potential (GAP) with
gapModelandpredict_gap()SchNetPack with
schnetModelandpredict_schnet()
Examples#
Prediction of (H2O)6 using mbGDML#
import numpy as np
from mbgdml.mbe import mbePredict
from mbgdml.models import gdmlModel
from mbgdml.predictors import predict_gdml
from mbgdml.descriptors import Criteria, com_distance_sum
from mbgdml.utils import get_entity_ids
# Loading mbGDML models.
model_paths = [
'./1h2o-model-train1000.npz',
'./2h2o-model-nbody-train1000.npz',
'./3h2o-model-nbody-train1000.npz'
]
model_comp_ids = [['h2o'], ['h2o', 'h2o'], ['h2o', 'h2o', 'h2o']]
model_desc_kwargs = (
{'entity_ids': get_entity_ids(atoms_per_mol=3, num_mol=1)},
{'entity_ids': get_entity_ids(atoms_per_mol=3, num_mol=2)},
{'entity_ids': get_entity_ids(atoms_per_mol=3, num_mol=3)},
)
model_desc_cutoffs = (None, 6.0, 10.0)
model_criteria = [
Criteria(com_distance_sum, desc_kwargs, cutoff) for desc_kwargs, cutoff \
in zip(model_desc_kwargs, model_desc_cutoffs)
]
models = [
gdmlModel(path, comp_ids=model_comp_id, criteria=criteria) \
for path, model_comp_id, criteria \
in zip(model_paths, model_comp_ids, model_criteria)
]
mbe_pred = mbePredict(models, predict_gdml)
# Structure information. This often comes from structure or data sets.
Z = np.array([8, 1, 1, 8, 1, 1, 8, 1, 1, 8, 1, 1, 8, 1, 1, 8, 1, 1])
R = np.array(
[[[-1.73521802, -1.13083385, 0.32487853],
[-1.54501802, -1.25583385, -0.62092147],
[-1.84191802, -0.15413385, 0.35947853],
[-1.43631802, 1.61886615, -0.08302147],
[-1.17431802, 1.32596615, -0.97352147],
[-0.58621802, 1.75866615, 0.37227853],
[-0.54571802, -0.22923385, -2.18532147],
[-0.48351802, -0.31643385, -3.14412147],
[ 0.38158198, -0.29733385, -1.85512147],
[ 1.87418198, -0.38073385, -0.90452147],
[ 1.98418198, 0.47796615, -0.46422147],
[ 1.65288198, -0.95933385, -0.15152147],
[ 0.63868198, -1.29043385, 1.52137853],
[-0.28361802, -1.33203385, 1.14077853],
[ 0.67688198, -1.97713385, 2.19787853],
[ 1.12828198, 1.42786615, 1.29217853],
[ 1.43688198, 1.95886615, 2.03657853],
[ 0.99038198, 0.52476615, 1.64897853]]]
)
entity_ids = np.array([0, 0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 5])
comp_ids = np.array(['h2o', 'h2o', 'h2o', 'h2o', 'h2o', 'h2o'])
# Predict total energies and forces.
E, F = mbe_pred.predict(Z, R, entity_ids, comp_ids)
print(E) # kcal/mol; shape: (1,)
# [-287373.68561825]
print(F) # kcal/(mol A); shape: (1, 18, 3)
"""
[[[ 1.88852751, 4.27617405, -3.10879603],
[-1.1754238, -0.56810535, 2.3096842 ],
[-0.88610451, -3.70003292, 0.68926709],
[ 4.77715706, -2.22894343, -2.95228678],
[-1.8784668, 1.9091819, 2.28471297],
[-2.2173002, 0.49008601, -0.02705656],
[ 4.90857346, -0.01682426, -1.80407224],
[-1.7842295, 0.53044132, 3.16420672],
[-3.08358393, -0.33648383, -0.37426009],
[-1.15415319, 1.8945924, 3.83795906],
[ 0.55584665, -2.03159006, -1.76731357],
[ 0.9244226, 0.22981244, -1.81504247],
[-4.80053866, -2.40925025, 0.9304219 ],
[ 3.6064161, 0.55564296, 0.93643006],
[ 1.13709823, 2.48598204, -2.15937521],
[ 0.72570156, -3.22025435, 3.45893275],
[-1.07780526, -0.27544762, -2.60668182],
[-0.46613732, 2.41501895, -0.99672996]]]
"""